Flowering plant evolution: systematics, speciation, natural history
Monarda brevis
Monarda eplingiana
Genomics and systematics
Monarda (Lamiaceae)
First reference genomes for Monarda: M. citriodora (diploid) and M. didyma (tetraploid).
Genus-wide whole genome sequencing for systematics and historical+contemporary reticulation.
Phlox (Polemoniaceae)
Association study + signatures of selection surrounding key floral color loci.
Demographic modeling for system of three shallowly diverged annual species.
Pedicularis (Orobanchaceae)
Biogeography of P. cranolopha in the Hengduan Mountains in Sichuan, China.
Methods and theory
Multispecies coalescent model + recombination
Simulation study showing influence of species tree on genealogical turnover along the genome.
Deriving waiting distance to genealogy change along the genome, given an arbitrary species tree.
Software tools
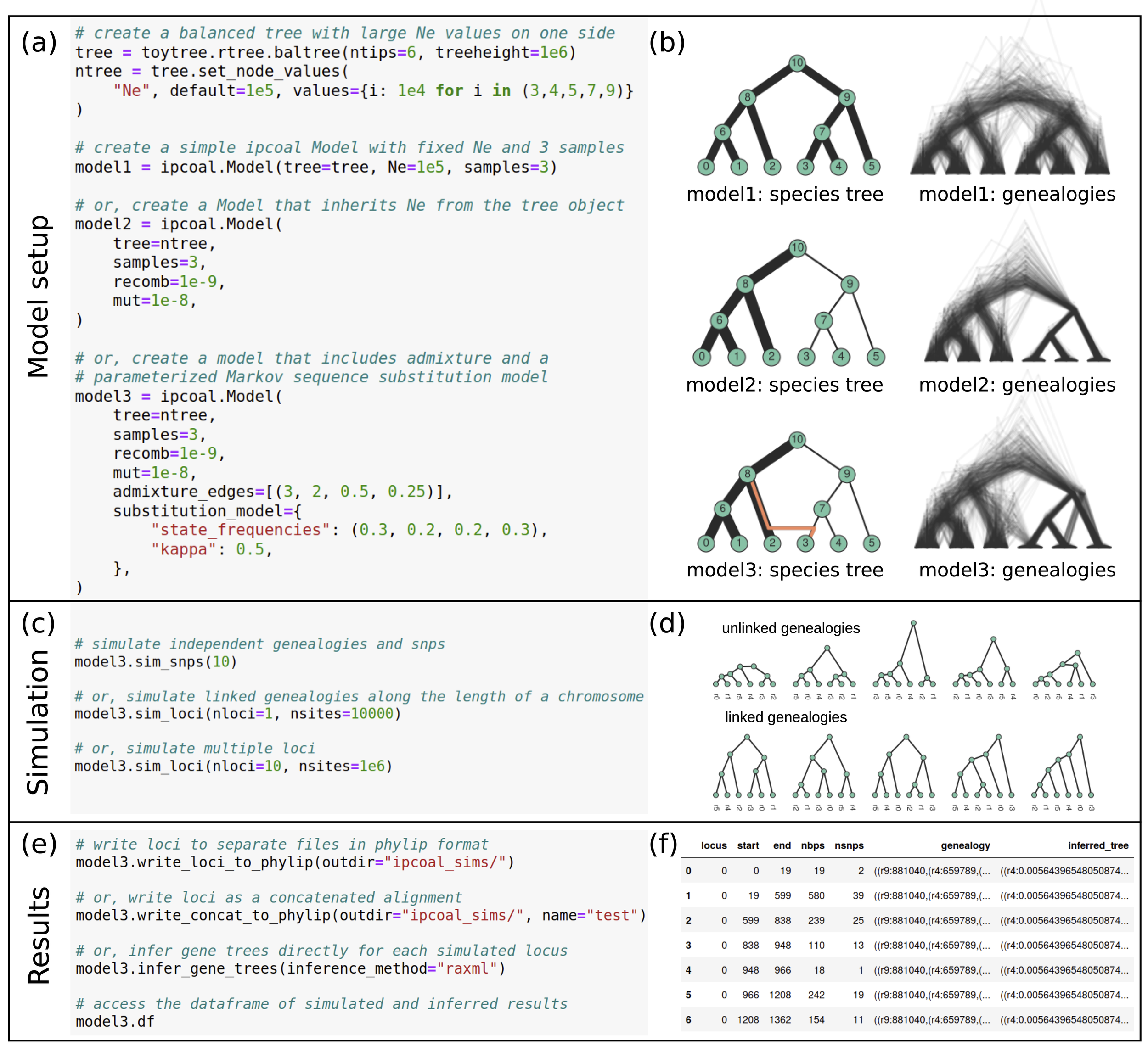
ipcoal: a python package enabling phylogenomic simulations
simcat: a machine-learning method to infer historical reticulation from SNP data.
SMC embedded in a species tree.
Demonstration ipcoal functions from our paper in Bioinformatics.
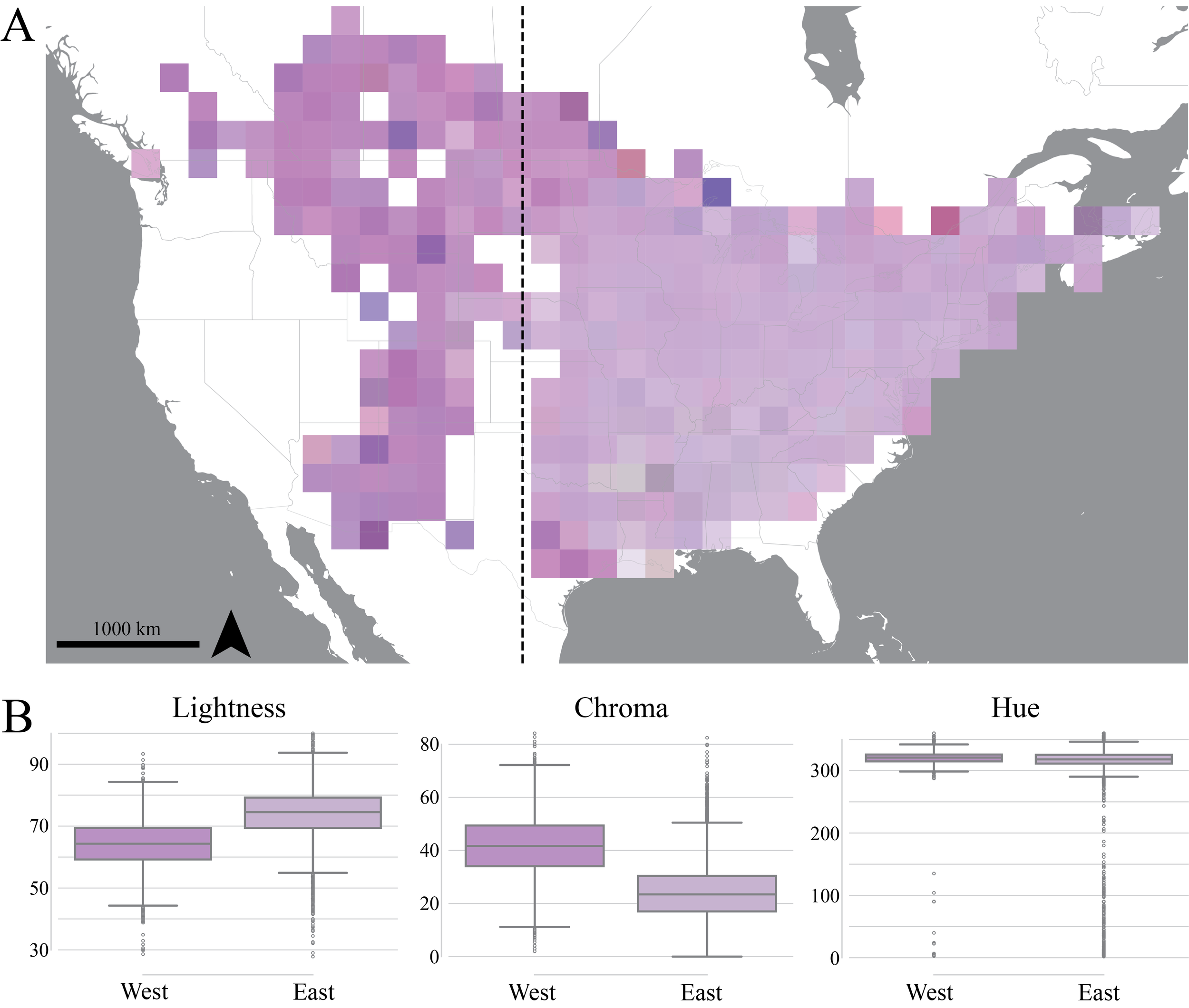
Inferring Monarda fistulosa flower color from iNaturalist data.
Red and orange flowers bloom later in the eastern U.S. in response to hummingbird migration.
Community science, natural history, taxonomy
Computational tools and community science data for large-scale natural history
Delayed flowering or red flowers in response to hummingbird migration
Flower color divergence in Monarda fistulosa
Taxonomy in Monarda and Phlox
Monarda mexicana (left) vs. two different specimens of M. citriodora var. austromontana.