New paper in Systematic Biology!
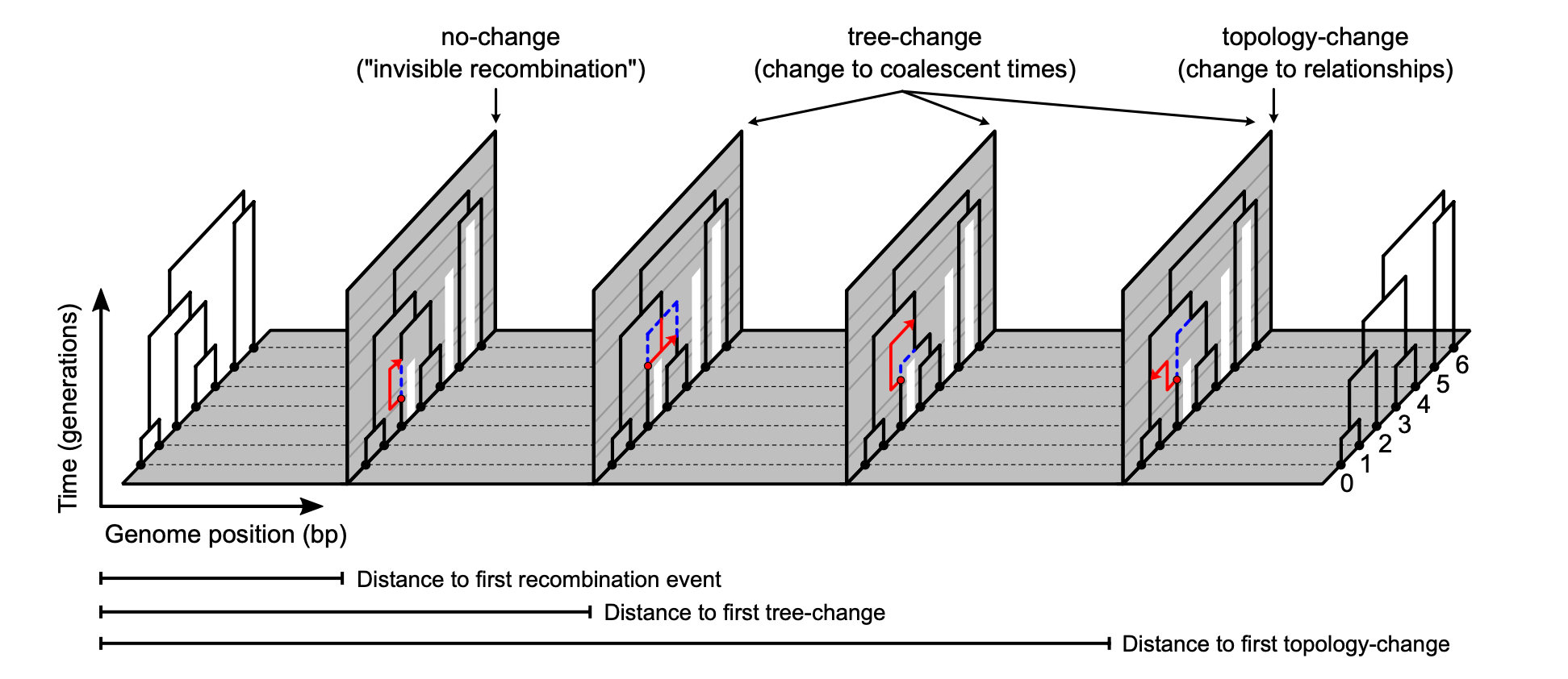
My new paper with Deren Eaton was just published in Systematic Biology! We derived distributions for -- given an arbitrary species tree model -- how far you have to move along a genome before observing a change in the underlying genealogy. Check it out here: https://doi.org/10.1093/sysbio/syaf059 [Estimating waiting distances between genealogy changes under a Multi-Species Extension of the Sequentially Markov Coalescent]
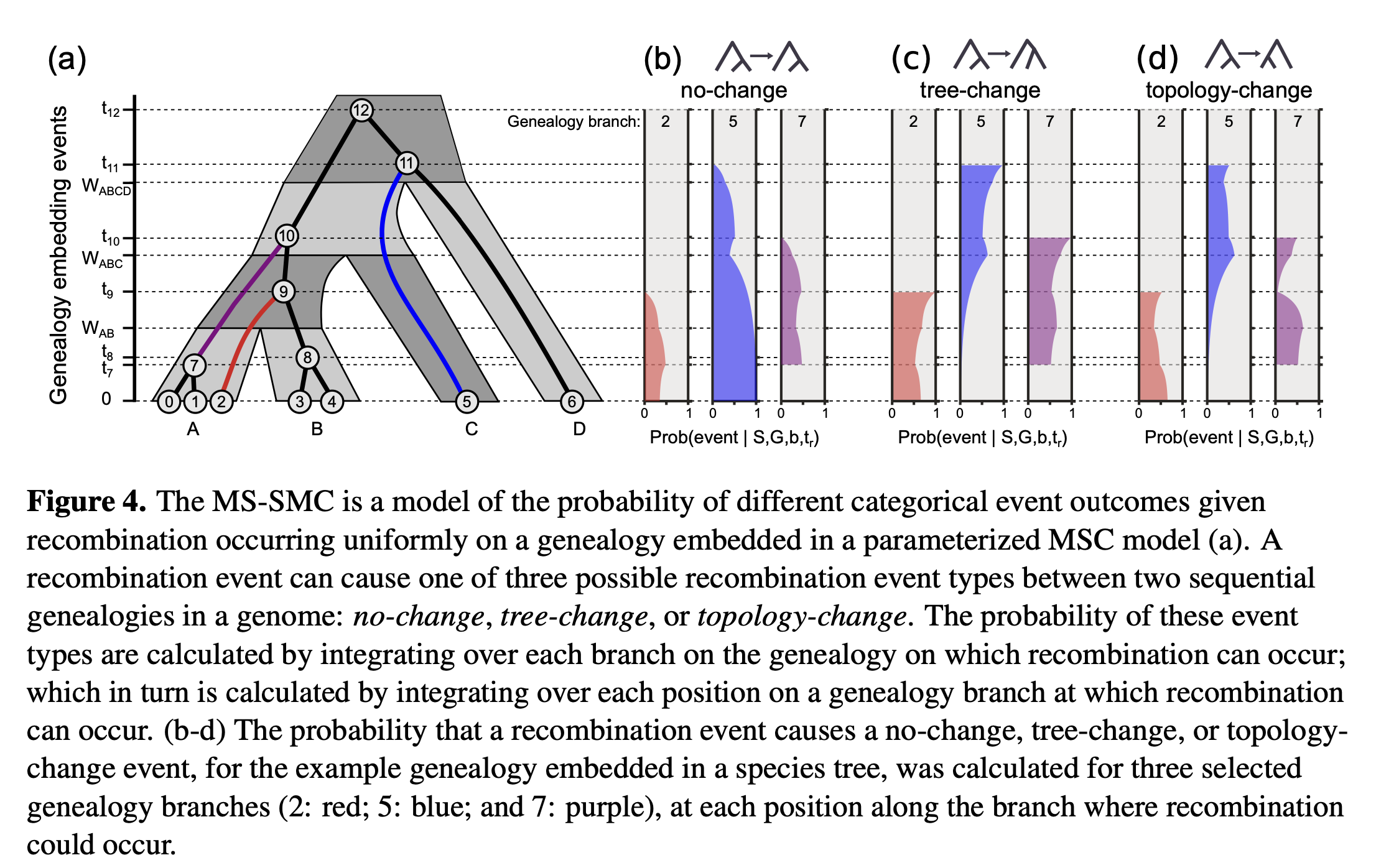
Alignments are mosaics of autocorrelated genealogies — nearby bases share ancestry until recombination alters the local tree. We derived the distributions of waiting distances for tree-change and topology-change events, extending prior single-population results to arbitrary species tree models. This explicitly links observable turnover in local ancestry to MSC model parameters for the first time!
We implement the math in ipcoal, and we validate against coalescent simulations. Finally, we demonstrate that there is information from observed waiting distances alone for fitting MSC parameters (Ne, divergence times, recombination rate).
There's value in leveraging linked genomic signals (vs. just unlinked gene trees) to learn about species history and demography. In the future we're hoping to see this plugged into species-tree inference and/or ARG inference pipelines to get more out of whole-genome data. Super grateful to colleagues and reviewers who greatly improved early versions of this manuscript. We're really proud of it.
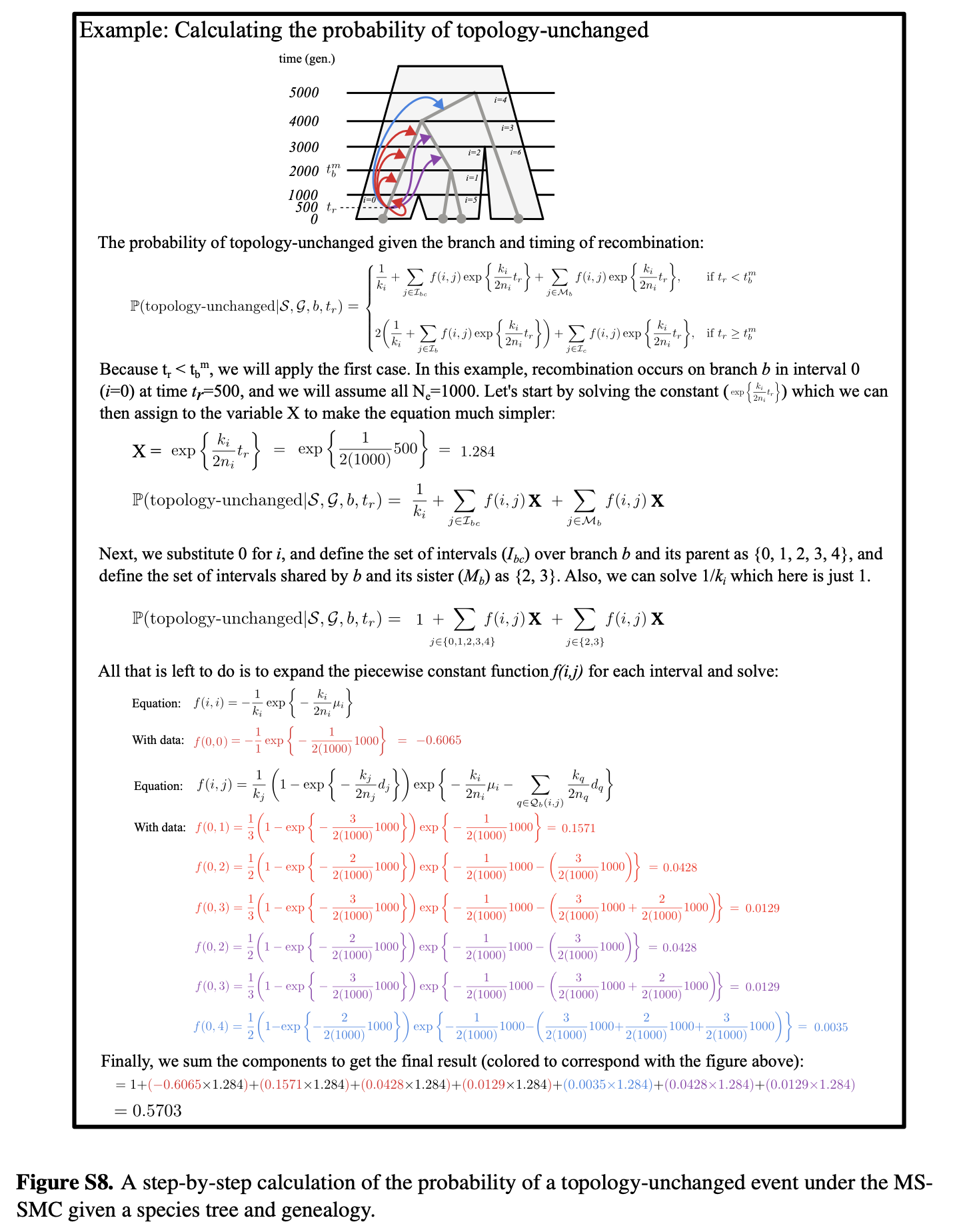
PS — There’s a lot of math in this paper, but we tried out best to make it accessible, including with a couple demonstrations of the calculations in the supplement: